About DNBSEQ
We’re redefining next-generation sequencing (NGS) capabilities and opening advanced applications so you can further your research. Our proprietary DNBSEQ technology provides extremely efficient and accurate real PCR-free NGS.
About DNBSEQ
We’re redefining next-generation sequencing (NGS) capabilities and opening advanced applications so you can further your research. Our proprietary DNBSEQ technology provides extremely efficient and accurate real PCR-free NGS.
How does DNBSEQ compare to
traditional methods?
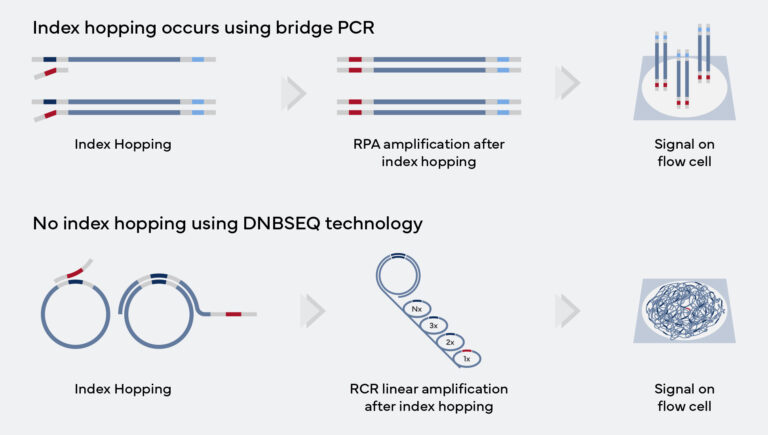
Higher Accuracy – Lower Cost – More Efficient
| Feature | DNBSEQ™ | PCR Clones |
|---|---|---|
| No clonal errors | ||
| Reduced indel errors | 3x less | |
| Suitable for Non-UMI applications | ||
| No index hopping | ||
| Allows ultra-dense patterned array | ||
| Supports ultra-higher throughput array | ||
| High occupancy on flow cell | ~95% | 60-70% |
| No qPCR | ||
| Low Duplication Rate | <2% | >2% |
See DNBSEQ in Action
DNBSEQ in Action
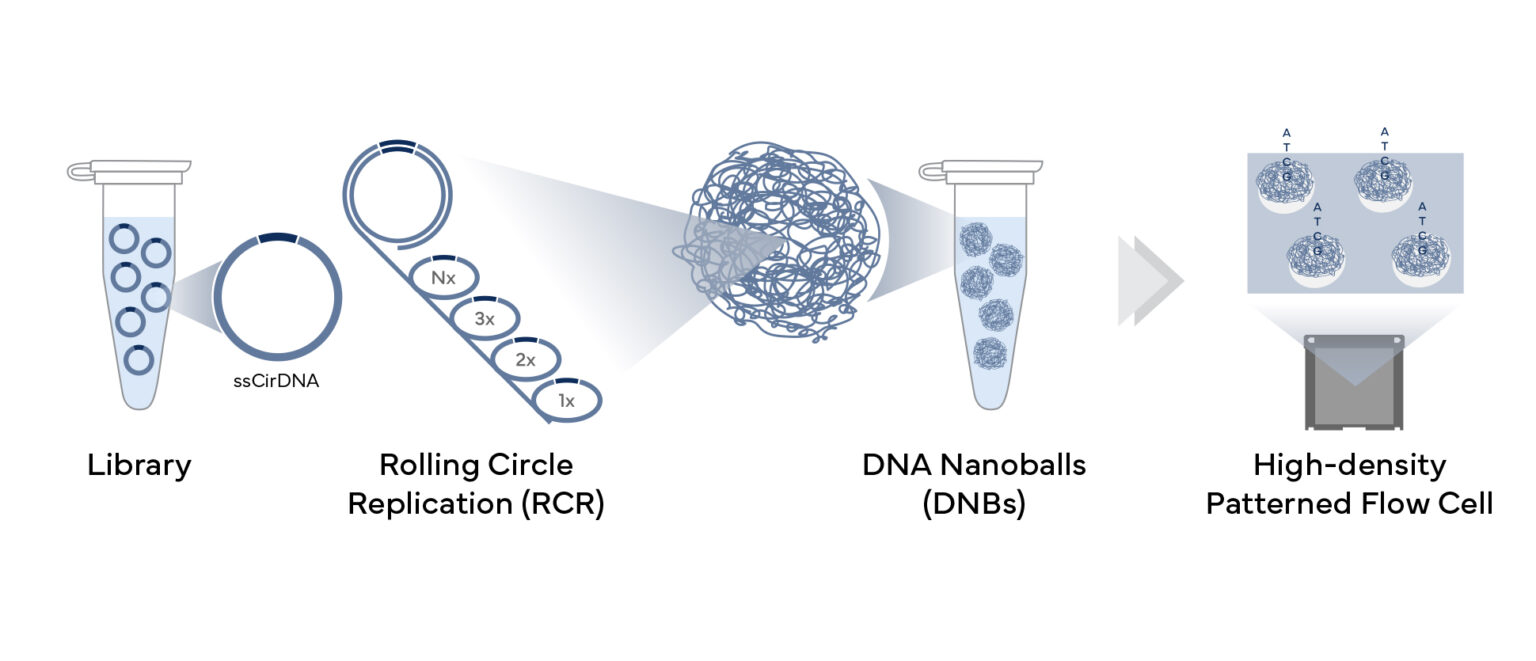
How Does Our Sequencing Technology Work?
Called DNBSEQ, our core technology is built around DNA nanoballs (DNBs). Created during library prep, compact DNBs are loaded onto flow cells and the sequence is read with four fluorescent probes recognizing four DNA bases. DNBSEQ then uses lasers to excite the probes, while taking millions of images to identify bases. These sharp and bright images are then fed into our proprietary image analysis algorithms to precisely sequence each sample.
Library Preparation
During sequencing library prep, the single-stranded circular DNA (ssCirDNA) molecules are created. Typical double-stranded DNA fragments, with adapter sequences at the terminal ends, are heated to generate ssDNA. A splint oligonucleotide is hybridized to both ssDNA ends to form nicked circles, and a DNA ligase repairs the nick to create complete single-stranded circles.
With the single-stranded circle as a template, DNBSEQ uses rolling circle replication (RCR) to create billions of DNA nanoballs (DNBs) in a single tube. Each copy is made from the original DNA circle, eliminating clonal amplification errors, and reducing the GC biases and dropouts, often produced by PCR. Under proprietary conditions, the concatemer with about 300-500 template copies is folded into a tiny DNA nanoball ~200nm in diameter.
Sequencing
The DNBs are loaded onto flow cells, which are etched with a pattern of uniformly-spaced ~200nm binding sites at submicron distances. Each site binds single DNB, ensuring high yield of accurate reads with sharp and bright signals and no interference from neighboring nanoballs.
The flow cell loading requires no expensive DNA quantification instruments or reagents. There is no under- or over-loading in a wide range of DNB concentrations. Proprietary loading buffers ensure DNBs stick to the same spots for hundreds of cycles, maintaining strong signals. These refinements enable exceptional sequencing accuracy with minimal reagent consumption.
From there, combinatorial probe-anchor synthesis (cPAS) chemistry hybridizes sequencing primers to the DNBs and fluorescently labeled reversibly terminated probes are incorporated by a proprietary DNA polymerase in consecutive sequencing cycles. The fluorescent probes are then excited by laser light and the DNB array is imaged using advanced cameras.
After completing the first strand, the second, complementary strand attached to the original DNB is synthesized by controlled MDA and sequenced. This generates a stronger second strand signal with high sequencing accuracy.
A Complete Solution for Every Step of Your Workflow

Kits & Reagents
No matter your workflow, our kits are simple to use and easily integrated. We also offer conversion kits for 3rd party library prep.

Lab Automation
With solutions from sample extraction to lab automation you can spend less time overseeing equipment and pursue more useful activities.


