Stereo-seq OMNI for FFPE Solution
Advance Spatial
Discovery in FFPE
Revolutionize Spatial Biology for Preserved FFPE Samples
Stereo-seq OMNI transcriptomics solution for formalin-fixed, paraffin-embedded (FFPE) tissue delivers true single-cell resolution and comprehensive RNA profiling. Designed to overcome the challenges of RNA degradation and protein cross-linking inherent to FFPE processing, the Stereo-seq OMNI solution utilizes an innovative Random Probe hybridization approach to capture total RNA, including coding, non-coding transcripts, as well as microbial RNAs efficiently.
Stereo-seq OMNI transcriptomics solution works efficiently in samples with RNA integrity levels as low as DV200 ≥ 30%, opening vast archives of clinically annotated tissues for spatial transcriptomics analysis. With its ability to work in diverse tissues from various species, the Stereo-seq OMNI transcriptomic solution for FFPE helps uncover relationships between gene expression, cellular morphology, and microenvironmental factors.
Total RNA Capture
Innovative Random Probe design enables efficient capture of coding and non-coding RNAs
For Low-Quality Tissues
Compatible with archived clinical specimens (DV200 > 30%)
True Single-Cell Resolution
Visualize gene expression patterns at sub-micron resolution with exceptional clarity
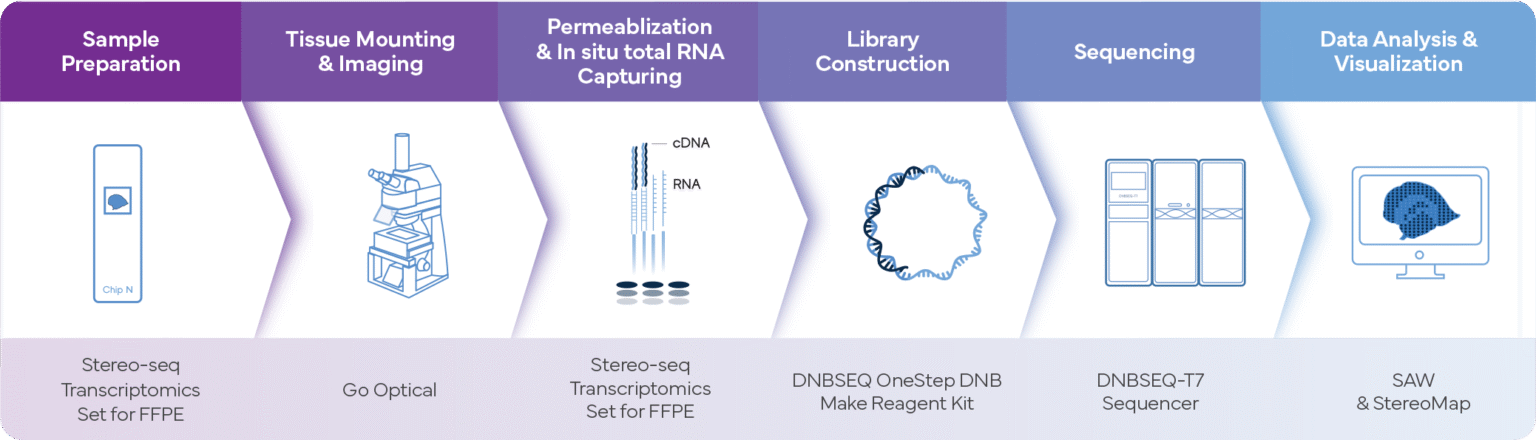
Complete Spatial Transcriptomics Workflow for FFPE
StereoMap is a desktop application designed to provide the essential analysis functionality you need to explore your Stereo-seq data interactively.
Applications
Stereo-seq OMNI transcriptomics solution for FFPE tissue integrates seamlessly with standard histopathology workflows, enabling correlative analysis of gene expression patterns with morphological features in the same tissue section. This capability is transformative for studying tumor heterogeneity, immune cell interactions, and developmental processes in archived samples, bridging historical clinical data with spatial insights.
Track Disease Progression
Stereo-seq OMNI transcriptomics solution for FFPE tissues identified distinct transcriptional disruptions across cortical layers in Alzheimer’s disease patients’ prefrontal cortex, highlighting significant reduction in SLC1A2-GRIA2 interactions closely linked to cognitive decline (Gong et al., 2025).

Stereo-seq OMNI transcriptomics solution’s nanoscale resolution and whole-transcriptome coverage allowed precise layer-specific pathological mapping, highlighting neuronal loss in layer II-VI, and significant glutamate signaling reduction in layer V.
Tumor Microenvironment Crosstalk
Stereo-seq OMNI transcriptomics solution for FFPE tissues identified spatially distinct transcriptional subclusters, demonstrating an enrichment of non coding RNAs like AL357507.1 at invasive tumor fronts (Ferri-Borgogno et al., 2024).

Stereo-seq OMNI transcriptomics solution identified novel biomarkers like stromal-specific SAMD4A, and tumor-stromal interface-specific CADPS, offering insights into the molecular basis of spatial cell heterogeneity and crosstalk within the tumor microenvironment.
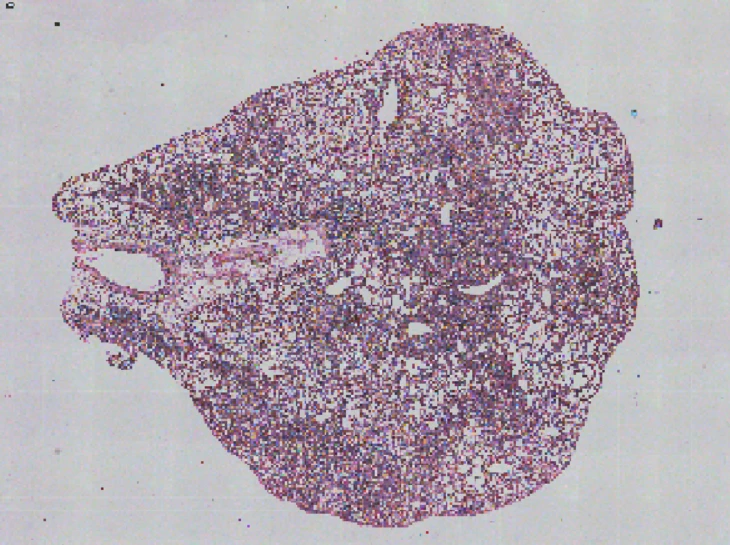
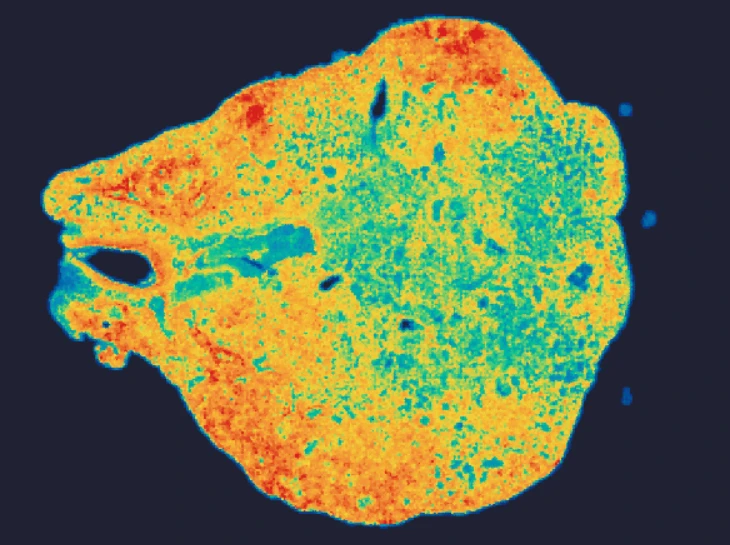
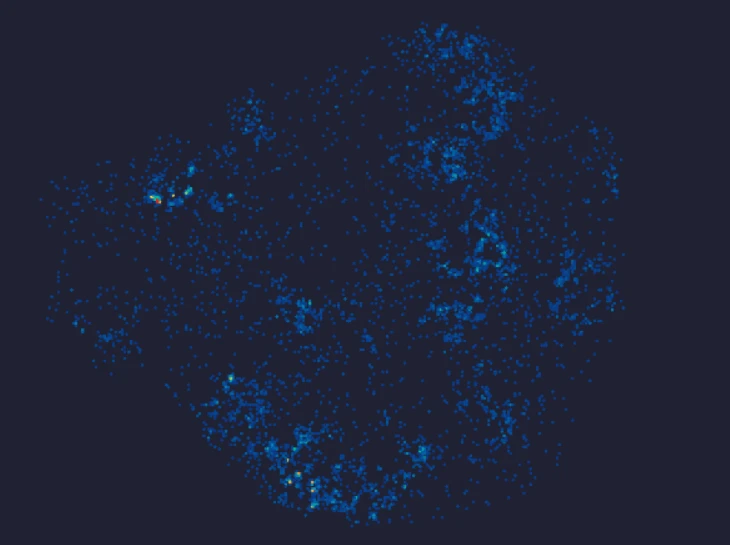
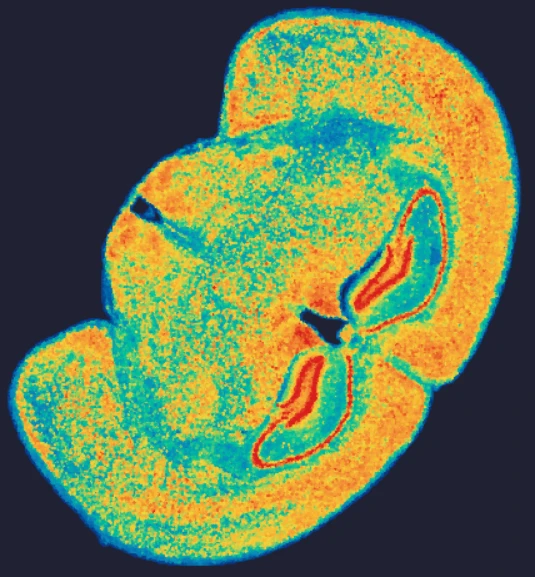
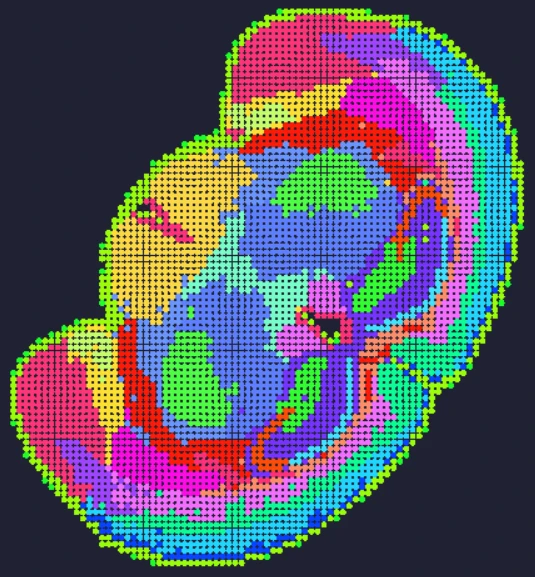
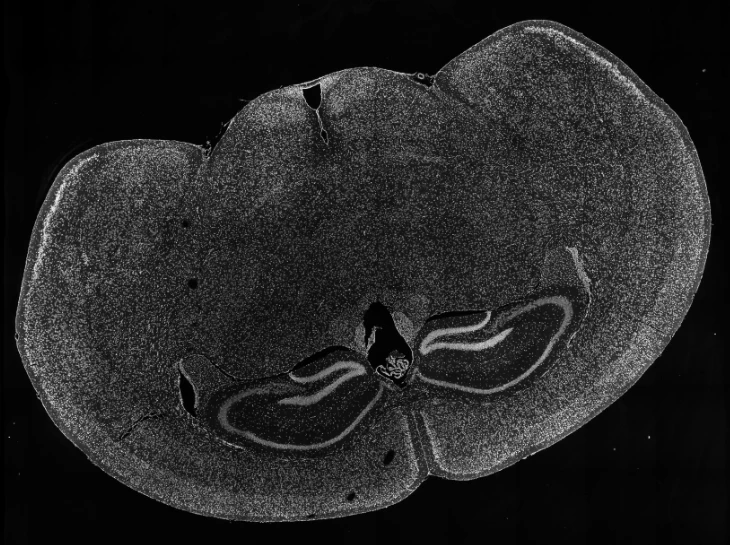
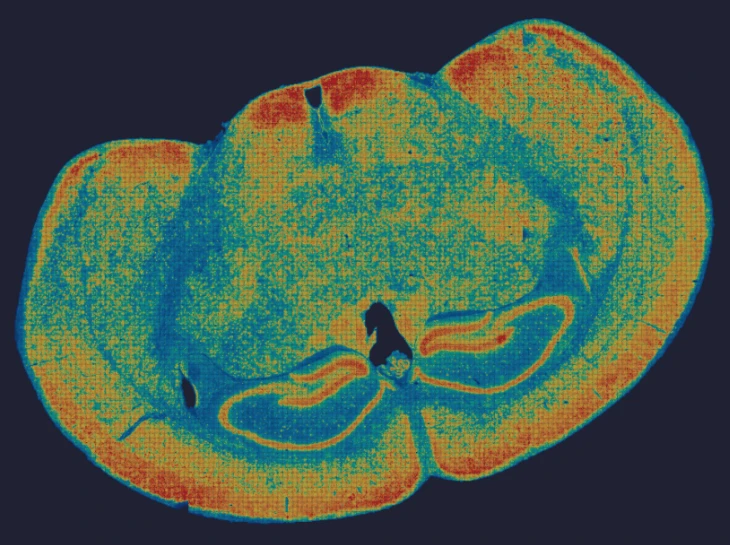
Demo Results
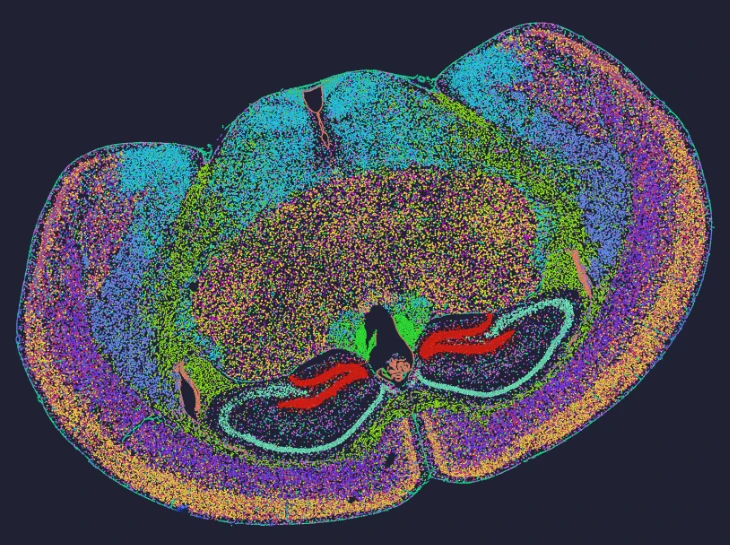
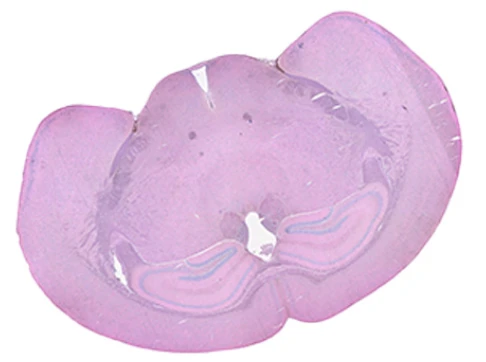
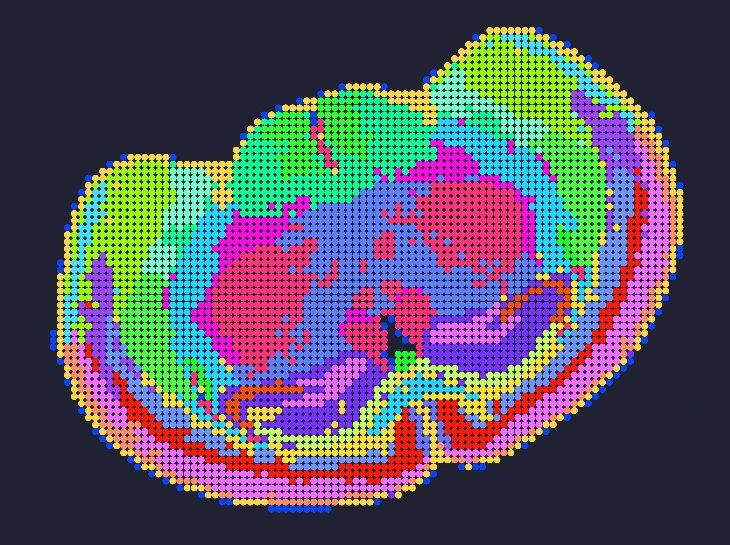
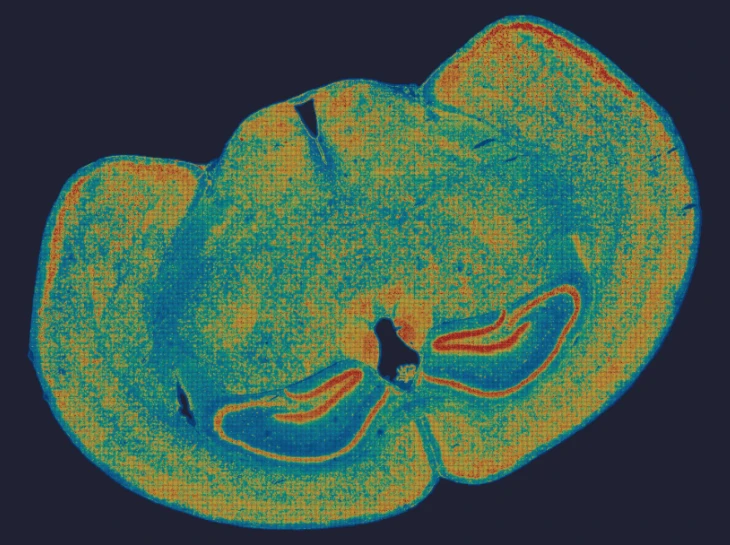
Exploring true spatial single-cell total RNA information with Stereo-seq OMNI
Ordering Information
| Category | Product | Cat. No. |
|---|---|---|
| Sample Prep | Stereo-seq OMNI Transcriptomics FFPE Set for Chip-on-a-slide (1cm*1cm) V1.1 | 211SN11114-CG |
| Stereo-seq OMNI Transcriptomics FFPE Set for Chip-on-a-slide (0.5cm*0.5cm) V1.1 | 211SN11004-CG | |
| Library Prep | Stereo-seq 16 Barcode Library Preparation Kit V1.1 | 111KL11160-CG |
| Instruments | STOmics Microscope – Go Optical | 900-001019-00 |
| Genetic Sequencer DNBSEQ-T7 | 900-000698-00 | |
| Sequencing Reagents | DNBSEQ-T7RS Stereo-seq Visualization Reagent Set (FCL PE75) | 940-001889-00 |
| DNBSEQ-G400RS Stereo-seq Visualization Reagent Set (FCL PE75) | 940-001885-00 |